Within the scope of molecular biology research, processing and analyzing DNA sequence files is a key daily task. A powerful and easy-to-use Mac tool can significantly improve efficiency.
Core functions and positioning
This tool is specially designed for processing DNA sequence files on macOS systems. It can support the creation, browsing and sharing of sequence files. These files are richly annotated and can be up to 1GB in size. This design solves the problems of inconsistent formats and data dispersion that often plague researchers when they integrate metadata such as gene annotations and mutation information.
There is such an integrated application whose target users are clearly molecular biologists, genetic researchers, and scholars in the field of bioinformatics. For professionals who frequently review large DNA sequence data, it simplifies multiple steps within an intuitive interface. For professionals who need to frequently compare large DNA sequence data, the above applications can also simplify operations accordingly. For professionals who need to frequently exchange large DNA sequence data with others, this application can also simplify the process and avoid the cumbersome switching between different professional software.
Compatible with a wide range of file formats
A variety of commonly used bioinformatics file formats can be opened by this application, which is an important advantage. Regardless of whether the data comes from public databases, sequencing companies, or cooperative laboratories, users can generally directly import and browse the data without tedious format conversion. This compatibility reduces data preprocessing time and allows researchers to enter the substantive analysis stage more quickly.
The ability to directly open files also lowers the technical threshold. Even graduate students or technicians who are not familiar with command line operations can use it. Data can be easily loaded through the graphical interface, and the composition, length, and related annotation information of the sequence can be visually viewed, making the preliminary review of DNA data more convenient.
Intuitive analysis and modification operations
In this application, in addition to viewing functions, it also allows users to perform basic analysis and modification operations on DNA profiles. Users can directly mark sequences on the graphical interface, highlight specific areas, or perform some basic editing actions. This provides convenience for quickly organizing data and preparing presentation materials.
The modification function does not refer to the direction of deep sequence reprogramming, but focuses on the addition, deletion, modification and query of annotation information. For example, users can add customized experimental notes to a gene sequence and mark specific single nucleotide polymorphism sites. These modifications can be directly saved in the file, which facilitates subsequent tracing and team collaboration.
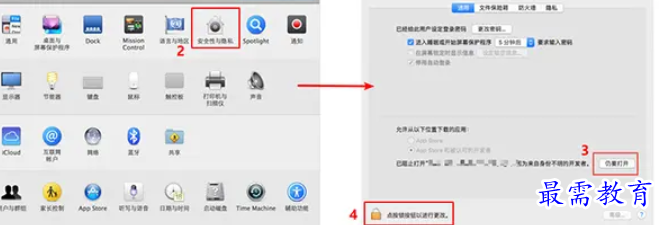
Dealing with Mac System Security Restrictions
In the relatively new macOS systems, especially after 10.12 Sierra, many applications downloaded from channels other than the App Store will encounter a "corrupted" prompt that cannot be opened. This is not a problem with the software itself, but rather the "Gatekeeper" security mechanism enabled by Apple by default that prevents applications developed by developers without official certification from running.
When encountering this situation, users can find and click "Open Anyway" in the "Security and Privacy" section of "System Preferences" to force start the application. The system will then remember this choice, which is a relatively quick and temporary solution.
Enable "Any Source" option
If the above method does not work for this situation, a more basic solution is to enable the installation permission selection such as "Any Source". Within the system range of macOS 10.12 to 10.14, users can find this option in the "Security and Privacy" settings area and manually check this action. This move enables users to run applications originating from any channel, including the Internet.
In macOS 10.15 Catalina and higher, this option is likely to be hidden by default. Users must open the Terminal application, enter specific command lines and instructions, and then re-enable it. After executing the command, enter the system preferences again and you will be able to see and select the "Any Source" option.
Share large data files
This tool's emphasis on sharing capabilities is critical for collaborative research. Users can directly share edited and annotated sequence files of up to 1GB with colleagues. Sharing may include generating a single project file containing all data and annotations, or sending it via email or cloud storage link.
This type of sharing ensures the integrity of the data. When the recipient uses the same software to open it, he can see the exact sequence view and all additional annotation information. This avoids format confusion or annotation loss caused by using different software to parse, thereby improving the efficiency of team communication and data reuse.
During your scientific research, which type of tool for processing DNA sequence data did you use most frequently? Is it based on graphical interface applications or command-line tools? Welcome to share your experiences and opinions in the comment area.



